It can be confusing to make sense of the alphabet soup of methods used to test if samples are actually part of an outbreak. In speaking with customers, we have heard a range of methods being used, from antibiograms without any further testing to custom SNP (Single Nucleotide Polymorphism) analysis that looks at each base of the genome. For those not deep into the technical nuances, the range of choices can be overwhelming. So, we thought a quick review could be helpful to place the methods they use in context.
Antibiograms & epidemiological surveillance are the traditional first pass methods for identifying possible healthcare associated infection (HAI) transmission. Although important and core to an investigation, they are never definitive. Relying only on surveillance makes it nearly impossible to confirm or rule-out transmission. While it is always good to put in precautionary measures when you suspect a possible transmission, high-stakes decisions or high-cost interventions benefit from knowing if you are dealing with a true transmission. What are the options available today and how did these technologies evolve?
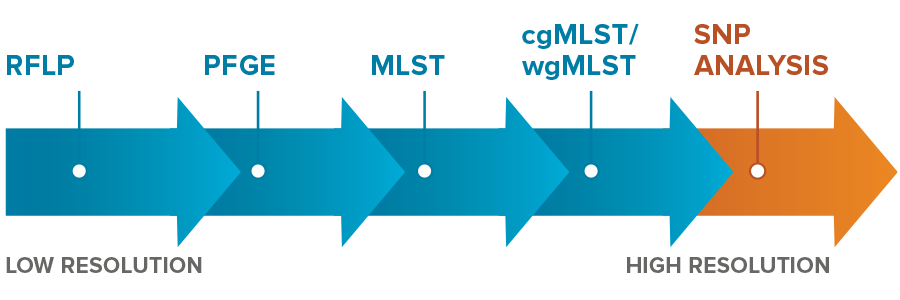
- RFLP (Restriction Fragment Length Polymorphism)—one of the original methods for genotypic investigation. We were surprised to find that RFLP is still used at some hospitals given its low resolution, inability to confirm clonality, and slow speed. More modern methods can be faster, cheaper, and more definitive.
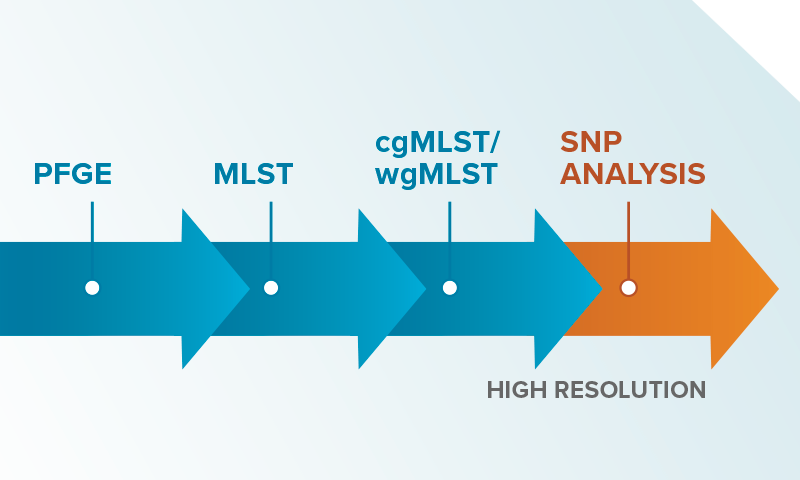
- PFGE (Pulse Field Gel Electrophoresis)—An older method of generating genomic information with limited resolution. Fewer and fewer places perform PFGE today because next generation sequencing can provide more accurate DNA sequences, cover far more of the genome, and is faster to execute. If you are still using PFGE, it’s time to upgrade.
- MLST (Multi-locus Sequence Type)—a genotypic method that looks at specific areas of the genome (<0.1% of the genome for many species) to group strains into types. MLST can be really good at telling you if two samples are part of a family, but it doesn’t tell you if they are twins. Although this may not be true in real life, in an HAI investigation, it’s the twins that cause all the trouble. In our projects, we often see samples of the same MLST that can be definitively ruled out as being clonal. We love it when that happens because it saves the hospital from the cost and time required for misdirected testing and unnecessary interventions.
- cgMLST/wgMLST (core- and whole-genome MLST)–uses far more of the genome to determine MLST type and offers much greater resolution. A very high resolution technique that has advantages for epidemiological surveillance, but that requires consensus schemes (common agreement on which loci are used for MLST categorization) which may not always be available.
- Whole-genome SNP analysis—uses differences at the single DNA base pair level to estimate genomic distance between samples. The highest resolution achievable but can be more difficult to execute well without sophisticated methods for dealing with complexities like plasmids and recombination events. It’s a good thing we have that. When we run samples through our process, we call SNPs relative to the closest matched reference genome (enabled by our unique ksim algorithm) as well as calling SNPs relative to a de novo assembly so that our results are sanity checked using a ‘belt and suspenders’ approach.
Let us know which one of these methods you are using. We would love to hear about your experiences and learn more about how HAI investigations are carried out in your hospital.
If it’s time for you to consider modernizing your HAI investigation process, contact us to learn more about epiXact®. epiXact uses high resolution, high depth whole-genome sequencing, combined with a SNP-based computational pipeline, to provide you with a simple, reliable answer about the relatedness of samples. We offer introductory service pricing that makes trying us out as easy and inexpensive as possible.